Reaction Parameters
This page lets you specify which parameters of the reaction you want to adjust.
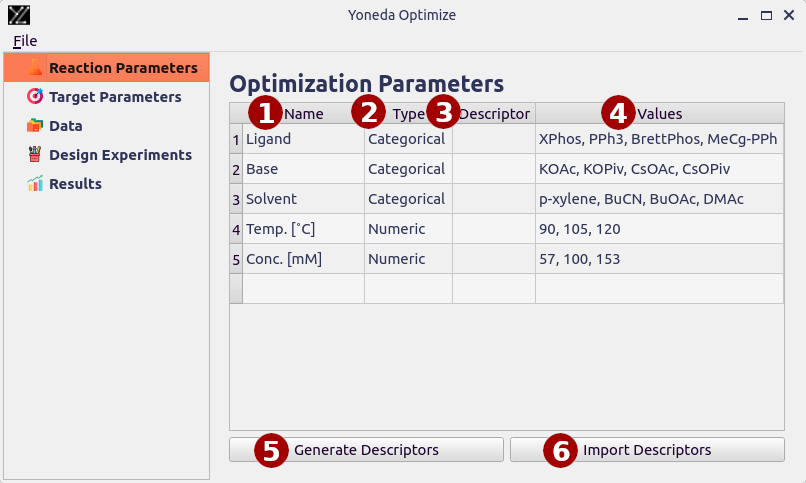
- Name
Name of the parameters. This can be any string, it is good practice to include units.
- Type
Either “Categorical” or “Numeric”, specifies whether the parameter should be treated as a number.
Examples of categorical parameters are solvents or ligands, numeric parameters include temperature or concentration.
- Descriptor
Name of the imported descriptors for this parameter, as show in the tree view on the left. Learn more about descriptors in the Chemical Descriptors section.
- Values
Allowed values for the parameter, separated by commas. You can also specify your range in the format
min:max|step_size. For example,0:100|10will generate values 0, 10, 20, …, 100. To ensure high-quality suggestions the total number of values should not be greater than 11.
- Generate Descriptors
This feature lets you generate chemical descriptors from SMILES.
- Import Descriptors
This lets you import custom chemical descriptors from a CSV file. The first column must contain the parameter values, the remaining columns will be treated as descriptors.
Note on numeric parameters
For best model performance, numeric parameters should be spaced roughly uniformly. If values of a parameter span multiple orders of magnitude, it is recommended to use logarithmic scale.
This can be best explained with examples:
Temperature: 90 ˚C, 105 ˚C, 120 ˚C |
✔ The values are spaced uniformly, 15 ˚C apart |
Substrate Equivalents: 0.8, 1.0, 1.5 |
✔ The values are spaced roughly uniformly, this is still fine |
Catalyst Equivalents: 0.01, 0.1, 1.0 |
✘ The values span multiple orders of magnitude, |
Log10 Catalyst Equivalents: -2, -1, 0 |
✔ The values are spaced uniformly, great solution! |